Serial dilution of bivalirudin for HPLC-MS-You can edit this template and create your own diagram. Creately diagrams can be exported and added to Word, PPT (powerpoint), Excel, Visio or any other document. Use PDF export for high quality prints and SVG export for large sharp images or embed your diagrams anywhere with the Creately viewer. It’s not enough to simply say “dilute before use on the skin”—this is insufficient information. You need to know which dilutions are safe and how to calculate them. With that information, you can use a chart like this to guide accurate dilution. DILUTION CHART Some numbers are rounded up or down to make measuring easier PIC’s dilution ratios are written as parts. Therefore, when PIC’s label suggest a dilution ratio of 1-to-4 (1:4) that means 1 part product and 4 parts water. Some people calculate dilution by dividing by 4 (in this example), which is an incorrect answer.
In some experiments it is pertinent to what the biologist is studying to ensure not only a proper colony count but that there is just one type of organism being studied. In previous blogs it is seen that these minuscule microorganisms stake their claim on whatever they see fit (so long as the right environmental factors comply) so how is it that biologists are able to separate them so that they may grow an individual microorganism and study it or even count it? That is easy (ok it’s rather time-consuming but worth all the effort) by serial dilution of course! Serial dilution is a series of dilutions, usually twofold or tenfold, used to determine the titer or concentration of a substance in a solution. Once an organism has been diluted out and allowed proper incubation time, this is when one can be counted. The way in which this experiment will ask you to count colonies will be by way of the naked eye. Plates that are suitable for counting should contain no less than 30 or no more than 300 colonies. In the below experiment you will be taken through just how to go about serial dilution of the specified microorganism and from there after proper incubation time be given the ratio on just how to count the number of colonies present.
What you will need:
*24-48 hour nutrient broth culture of Escherichia coli, six 20 mL nutrient agar deep tubes and seven sterile 9 mL water blanks.
*Hot plate, water bath, thermometer, test tube rack, Bunsen burner, sterile 1 mL serological pipette, mechanical pipetting device, sterile Petri dishes.
*Disinfectant solution in a 500 mL beaker, glassware marking pencil, bent glass rod, and beaker with 95% alcohol.
Procedure:
- Liquify six agar deep tubes by boiling in a water bath. Cool the molten agar tubes and maintain in the water bath at 45 degrees Celsius.
- Label the E. coli culture tube with the number 1 and the seven 9 mL water blanks as numbers 2 through 8. Place the labeled tubes in a test tube rack. Label the Petri dishes 1A, 1B, 2A, 2B, 3A, and 3B.
- Mix the E. coli culture (Tube 1) by rolling the tube between the palms of your hands to ensure even dispersal of cells in the culture.
- With a sterile pipette, aseptically transfer 1 mL from the bacterial suspension, Tube 1 to water blank Tube 2. Discard the pipette in the beaker of disinfectant. The culture has been diluted 10 times (1-10 dilution).
- Mix Tube 2 and with a fresh pipette, transfer 1 mL to Tube 3. Discard the pipette. The culture has been diluted 100 times (1-100 dilution).
- Mix Tube 3 and, with a fresh pipette, transfer 1 mL to Tube 4. Discard the pipette. The culture has been diluted 1000 times (1-100 dilution).
- Mix Tube 4 and, with a fresh pipette, transfer 1 mL to Tube 5. Discard the pipette. The culture has been diluted 10,000 times (1-10,000 dilution).
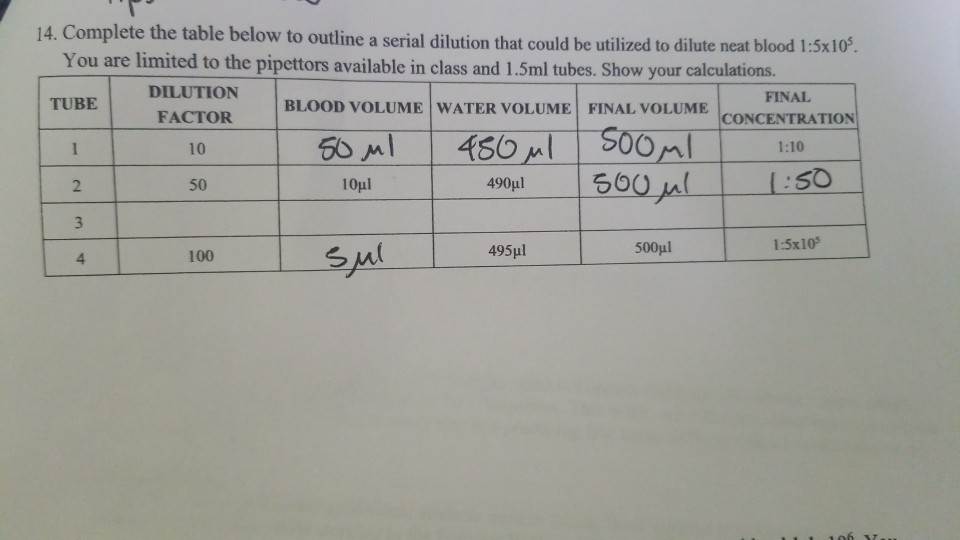
- Mix Tube 5 and, with a fresh pipette, transfer 0.1 mL of this suspension to Plate 1A. Return the pipette to Tube 5 and transfer 1 mL to tube 6. Discard the pipette. The culture has been diluted 100,000 times (1-100,000 dilution).
- Mix Tube 6 and, with a fresh pipette, transfer 1 mL of this suspension to Plate 1B. Return the pipette to Tube 6 and transfer 0.1 mL to Plate 2A. Return the pipette to Tube 6 and transfer 1 mL to Tube 7. Discard the pipette. The culture has been diluted 1,000,000 times (1-1,000,000 dilution).
- Mix Tube 7 and, with a fresh pipette, transfer 1 mL of this suspension to Plate 2B. Return the pipette to Tube 7 and transfer 0.1mL to Plate 3A. Return the pipette to Tube 7 and transfer 1 mL to Tube 8. Discard the pipette. The culture has been diluted 10,000,000 times (1-10,000,000).
- Mix Tube 8 and, with a fresh pipette, transfer 1 mL of this suspension to Plate 3B. Discard the pipette. The dilution procedure is now complete.
- Check the temperature of the molten agar medium to be sure the temperature is 45 degrees Celsius. Remove a tube from the water bath and wipe the outside surface dry with a paper towel. Using the pour-plate technique, pour the agar into Plate 1A rotating the plate gently to ensure uniform distribution of the cells in the medium.
- Repeat Step 12 for the addition of molten nutrient agar to Plates 1B, 2A, 2B, 3A and 3B.
- Once the agar has solidified, incubate the plates in an inverted position for 48 hours at 37 degrees Celsius.
After proper incubation time, proceed with the following:
- Observe all colonies on plates and count each colony. Statistically valid plate counts are only obtained from bacterial cell dilutions that yield between 30 and 300 colonies. Plates with more that 300 colonies cannot be counted and are designated as “too numerous to count-TNTC”; plates with fewer than 30 colonies are designated as “too few to count-TFTC.” Count only plates containing between 30 and 300 colonies. Remember to count all subsurface as well as surface colonies.
- The number of organisms per mL of original culture is calculated by multiplying the number of colonies counted by the dilution factor:
Number of cells per mL=number of colonies X dilution factor
Results:
The results of the serial dilution – agar plate analysis are given in the table below:
| Plate | Dilution Factor | DilutionPlated | Final dilution on plate | No. of colonies | Bacterial count1 | Ave. count2 |
| 1A | 105 | 0.1 mL | 10-4 | 175 | 1.75 x 106 | 0.006 |
| 1B | 105 | 1.0 mL | 10-5 | TNTC | TNTC | TNTC |
| 2A | 106 | 0.1 mL | 10-5 | 50 | 5.00 x 106 | 0.002 |
| 2B | 106 | 1.0 mL | 10-6 | TNTC | TNTC | TNTC |
| 3A | 107 | 0.1 mL | 10-6 | 1 | 1.00 x 109 | 0.1 |
| 3B | 107 | 1.0 mL | 10-7 | TNTC | TNTC | TNTC |
1 Bacterial count per ml of sample (CFU/mL)
2 Average count per mL of sample (CFU/mL)
Colony numbers “too numerous to count” are designated TNTC
There is some agreement between these results and the outcomes expected with a successful serial dilution regimen. With each dilution, a reduced number of colonies was observed (175, 50, and 1 for plates 1A, 2A, and 3A, respectively). The reduced counts between plate’s 1A and 2A were expected, and consistent with the regimen. Plate 3A produced only 1 colony, which actual renders it “to few to count”. The numbers of colonies on plates 1B, 2B, and 3B were “too many to count” despite the dilution.
The numbers of colony forming units was determined using the following equation:
Number of cells/ml = number of colonies x dilution factor
Those counts are given in the table above.
Since the duplications plated are replicates of each other, it was determined that the averages of the duplicate bacterial counts per ml of sample. These results are listed in Table 1 above. Since several plates produced colonies “too many to count”, the resulting averages are probably meaningless.
Conclusion:
The ability to determine with any reliability the numbers of viable organisms growing on the plates was not achieved in this experiment. Serial dilution is a method intended to reduce the numbers of colonies to a range between 30 and 300; two (2) of the six (6) plates we produced exhibited counts within the targeted range. (It should be noted that some level of dilution was achieved as demonstrated with colony counts of 175 and 50 on plate’s 1A and 2A, respectively.)
The failure to achieve dilution is probably results from incorrect technique (measurements or calculations). Inoculation and incubation were achieved as evidenced by colonies “too numerous to count.”


March 30, 2014 @ 7:15 p.m.
This section is not a recipe for your experiment. It explains someprinciples for designing dilutions that give optimal results. Onceyou understand these principles, you will be better able to designthe dilutions you need for each specific case.
Often in experimental work, you need to cover a range ofconcentrations, so you need to make a bunch of differentdilutions. For example, you need to do such dilutions of thestandard IgG to make the standard curve in ELISA, and then againfor the unknown samples in ELISA.
You might think it would be good to dilute 1/2, 1/3, 1/10, 1/100.These seem like nice numbers. There are two problems with this series ofdilutions.
- The dilutions are unnecessarily complicated to make. You need to do a differentcalculation, and measure different volumes, for each one. It takes a longtime, and it is too easy to make a mistake.
- The dilutions cover the range from 1/2 to 1/100 unevenly.In fact, the 1/2 vs. 1/3 dilutions differ by only 1.5-fold in concentration,while the 1/10 vs. 1/100 dilutions differ by ten-fold. If you are going tomeasure results for four dilutions, it is a waste of time and materialsto make two of them almost the same. And what if the half-maximal signaloccurs between 1/10 and 1/100? You won't be able to tell exactly where itis because of the big space between those two.
Serial dilutions are much easier to make and they cover the range evenly.
Serial dilutions are made by making the same dilution step over and over,using the previous dilution as the input to the next dilution in each step.Since the dilution-fold is the same in each step, the dilutionsare a geometric series (constant ratio between any adjacent dilutions).For example:
Serial Dilution Table
- 1/3, 1/9, 1/27, 1/81
- 1/5, 1/25, 1/125, 1/625
When you need to cover several factors of ten (several 'orders of magnitude') witha series of dilutions, it usually makes the most sense to plot the dilutions(relative concentrations) on a logarithmic scale. This avoids bunching mostof the points up at one end and having just the last point way fardown the scale.
Before making serial dilutions, you need to make rough estimatesof the concentrationsin your unknowns, and your uncertainty in those estimates. For example,if A280 says you have 7.0 mg total protein/ml, and you thinkthe protein could be anywhere between 10% and 100% pure, then yourassay needs to be able to see anything between 0.7 and 7 mg/ml.That means you need to cover a ten-fold range of dilutions, or maybe a bitmore to be sure.
If the half-max of your assay occurs atabout 0.5mg/ml,then your minimum dilution fold is(700mg/ml)/(0.5mg/ml) = 1,400.Your maximum is(7000mg/ml)/(0.5mg/ml) = 14,000.So to be safe, you might want to cover 1,000 through 20,000.
In general, before designing a dilution series, you need to decide:
- What are the lowest and highest concentrations (or dilutions)you need to test in order to be certain of finding the half-max? Thesedetermine the range of the dilution series.
- How many tests do you want to make? This determines the size of theexperiment, and how much of your reagents you consume. More tests will coverthe range in more detail, but may take too long to perform (or cost too much).Fewer tests are easier to do, but may not cover the range in enough detailto get an accurate result.
- What volume of each dilution do you need to make in order to haveenough for the replicate tests you plan to do?
Now suppose you decide that six tests will be adequate (perhapseach in quadruplicate).Well, starting at 1/1,000, you need five equal dilution steps (giving yousix total dilutions counting the starting 1/1,000) that end ina 20-fold higher dilution (giving 1/20,000). You can decide on a goodstep size easily by trial and error. Would 2-fold work? 1/2, 1/4, 1/8, 1/16, 1/32. Yes, in factthat covers 32-fold, more than the 20-fold range we need. (The exact answeris the 5th root of 20, which your calculator will tell you is 1.82 foldper step. It is much easier to go with 2-fold dilutions and gives about thesame result.)
How To Serial Dilution
So, you need to make a 1/1,000 dilution to start with. Then you need toserially dilute that 2-fold per step in five steps. You could make 1/1,000 byadding 1 microliter of sample to 0.999 ml diluent. Why is that a poor choice?Because you can't measure 1 microliter (or even 10 microliters) accuratelywith ordinary pipeters. So, make three serial 1/10 dilutions(0.1 ml [100 microliters] into 0.9 ml): 1/10 x 1/10 x 1/10 = 1/1,000.
Serial Dilution Steps
Now you could add 1.0 ml of the starting 1/1,000 dilution to1.0 ml of diluent, making a 2-fold dilution (giving 1/2,000).Then remove 1.0 ml from that dilution (leaving 1.0 ml for yourtests), and add it to 1.0 ml of diluent in the next tube (giving1/4,000). And so forth for 3 more serial dilution steps (giving1/8,000, 1/16,000, and 1/32,000). You end up with 1.0 ml of each dilution.If that is enough to perform all of your tests, this dilution planwill work. If you need larger volumes, increase the volumes you useto make your dilutions (e.g. 2.0 ml + 2.0 ml in each step).